Building a High-Recall Toxicity Screener
A journal of pivots: Solving data desyncs, disregarding theoretical metrics, and identifying structural blind spots in Graph Neural Networks.
00 // The Problem: The Cost of a Miss
In pharmaceutical auditing, a False Positive is an annoyance (a safe drug is flagged for review), but a False Negative is a disaster (a toxic drug is marked safe). Standard machine learning models optimize for overall accuracy, often sacrificing safety by missing rare but deadly edge cases.
The Engineering Objective: Architect a "High-Recall" Graph Neural Network (GNN) that aggressively flags potential toxins. The goal was to push Recall to >0.95 to ensure zero-tolerance for missed toxins, while integrating "White-Box" explainability (Atomic Heatmaps) to justify every flag to human auditors.
01 // The Mathematical Core
The screener utilizes neighborhood aggregation to update atomic feature vectors based on local chemical environments:
Spectral normalization ensures stable updates across different molecular sizes:
Final fixed-size representation is achieved by concatenating Global Mean and Max pooling to capture both general structural properties and localized "hotspots":
Mathematical Legend
Node-Level (Eq 1)
- x_i^{(l)} : Feature vector of atom i at layer l.
- σ : Non-linear activation function (ReLU).
- Θ : Learnable filter parameters (weights).
- 𝒩(i) : Neighbors of atom i (including self-loop).
- d̂ : Degree of node (used for normalization).
Matrix & Graph (Eq 2 & 3)
- Ã : Adjacency matrix with self-loops (A + I).
- D̃ : Diagonal degree matrix of Ã.
- V : Set of all atoms in the molecule.
- || : Concatenation (joining Mean & Max pools).
- X_{graph} : Final fixed-length molecular embedding.
02 // Data Acquisition & Integrity
Establishing a reproducible pipeline began with automated data acquisition. I implemented a script to download the official Tox21 dataset and verify the raw CSV structures before processing.
03 // Solving the Metadata Desync
Standard GNN pipelines often lose track of molecular SMILES strings during batching. I re-engineered the ingestion pipeline to attach metadata directly to the graph objects. This ensures every prediction—success or failure—is traceable.
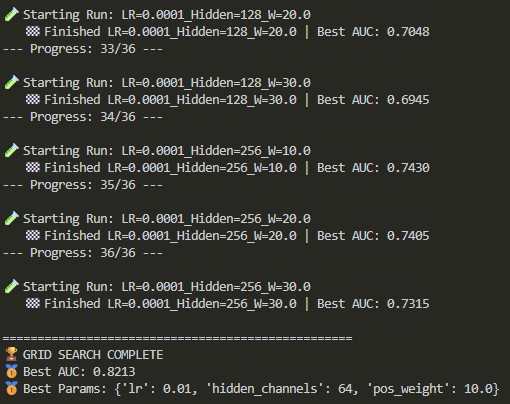
04 // 36-Model Grid Search
I executed a sweep across 36 unique configurations to identify the optimal baseline for Hidden Channels and initial weights.
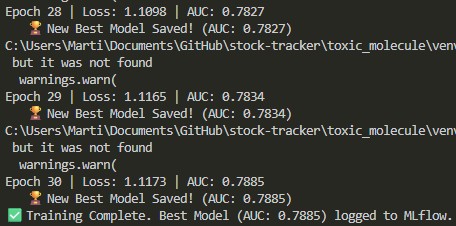
05 // Training: Precision through Optimization
Initially, a learning rate of \(0.01\) caused the model to oscillate, requiring a massive pos_weight to catch toxic signals. By slowing the learning rate to \(0.001\), the model achieved smoother convergence—correctly flagging toxic Dinitrotoluene while identifying Aspirin as safe using a standard \(10.0\) weight.
06 // Theoretical PR-Curves vs. Safety Audits
Theoretical F1 points on a PR curve often ignore the cost of missed toxins. I disregarded theoretical "optimums" for a raw Threshold Analysis.
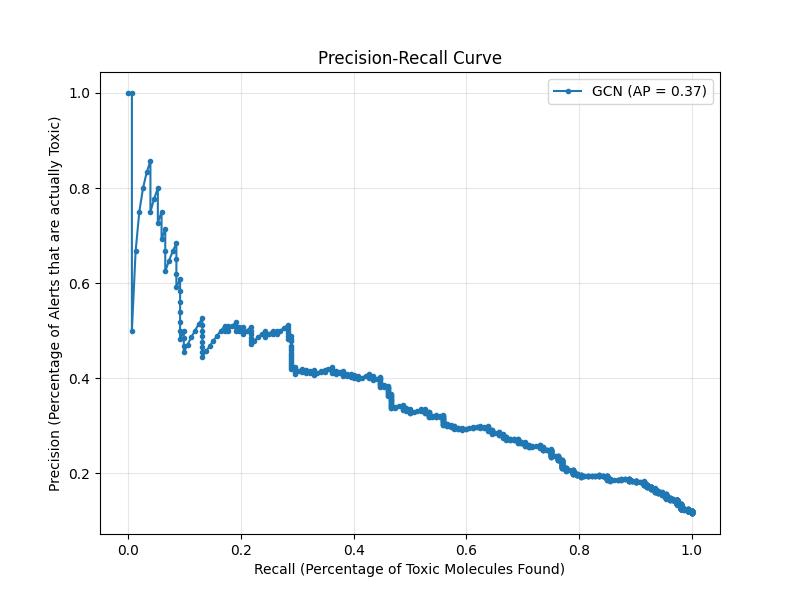
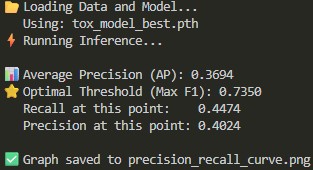

Selecting a 0.30 threshold secured a 0.9600 Recall.
07 // Explainable Ray Serve API
The model is deployed via Ray Serve to handle REST requests at scale. I integrated Captum's Integrated Gradients to provide atomic-level heatmaps for every prediction.
I then implemented a batch testing script to verify production performance and log real-time inference results.
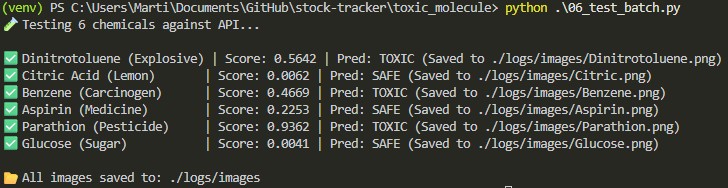

Batch Testing Case Studies
True Positives (Correctly Flagged)
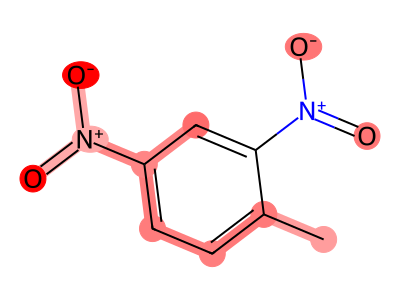
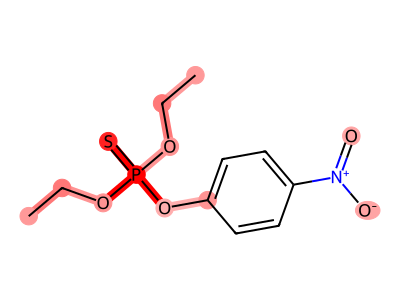
True Negatives (Correctly Classified Safe)
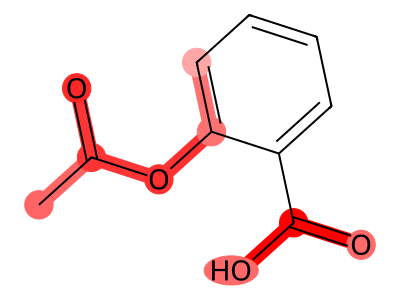
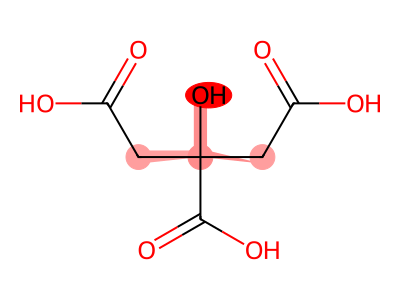
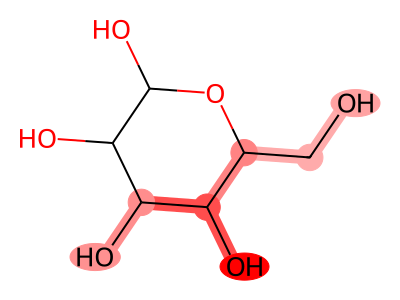
08 // Identifying the Topological Floor
Audit scripts identified a "Topological Floor" where structural signals wash out in simple graphs (7-15 atoms).
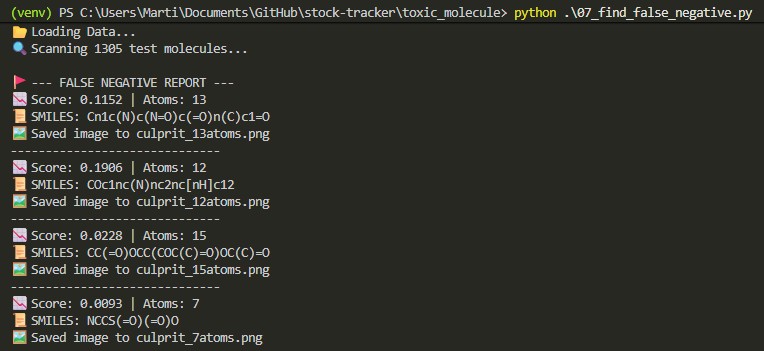
False Negative Gallery
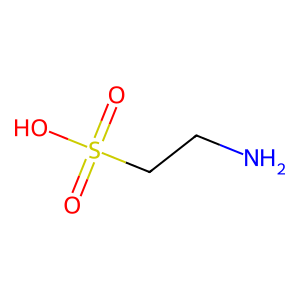
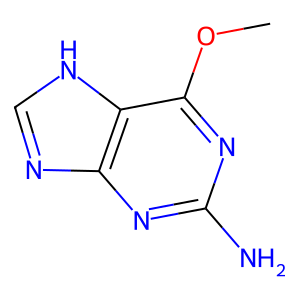
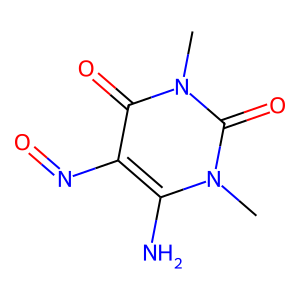
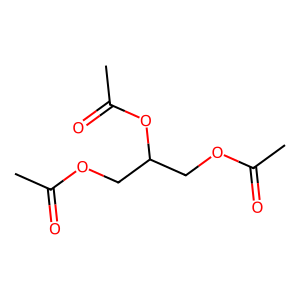
09 // Model Limitations & Future Outlook
- Subset Training Constraints: The current weights were optimized on a specific subset of the Tox21 dataset.
- The Arsenic Blind Spot: Rare elements like Arsenic cause confidence gaps due to feature underrepresentation.
- False Negative Management: Future models must integrate global molecular descriptors to assist the topological GNN.